This tutorial demonstrates how to use the vlMolecule module classes to manipulate and render molecules and chemical structures.
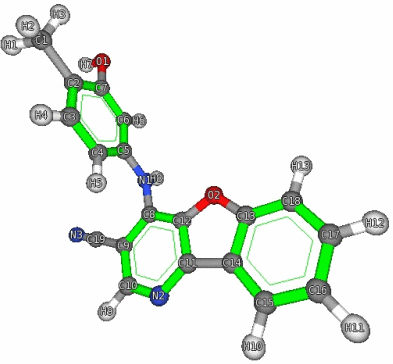
The vlMolecule module is a simple and minimalistic framework to manipulate and render chemical structures. The vl::Molecule class is the most important member of the module and represents a molecular structure as a collection of vl::Atom[s] and vl::Bond[s]. The vl::Molecule class allows you to add and remove both atoms and bonds in order to create any kind of molecule. The vlMolecule framework is flexible and intuitive, it allows you for example to set the color, the size, the visibility etc. for each atom and for each bond, it allows you to define the geometrical detail for bonds and atoms, it allows you highlight aromatic rings and much more.
The vlMolecule module also provide a comprehensive atom database, in fact using the vl::atomInfo() function the user can retrieve information like element name, element symbol, atom number, electronegativity, cpk color, various radii etc. for a given atom type (vl::EAtomType).
Using the vl::loadMOL2() function you can also load a MOL2 (or multi-MOL2) file.
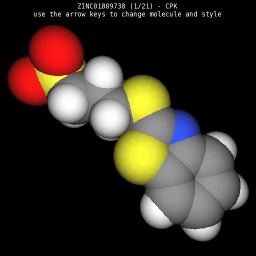
The example below implements a simple molecule viewer.
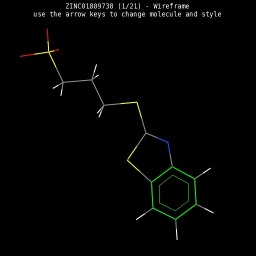
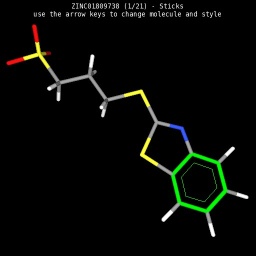
| 
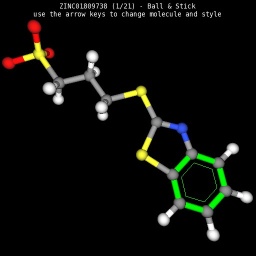
| 
| 
|
| Wireframe style | Sticks style | Ball & Sticks style | CPK space fill style |
[From App_Molecules.cpp]
class App_Molecules: public BaseDemo { public: App_Molecules(): mCurrentMolecule(0), mCurrentStyle(0), mText( new vl::Text ) {} void updateMolecule() { if (mCurrentStyle == 0) // wireframe { // activate "wireframe" style mMolecules[mCurrentMolecule]->setMoleculeStyle(vl::MS_Wireframe); // colorize the atoms by their CPK color mMolecules[mCurrentMolecule]->setCPKAtomColors(); // define the line width mMolecules[mCurrentMolecule]->setLineWidth(2.0f); // activate line anti-aliasing mMolecules[mCurrentMolecule]->setSmoothLines(true); } else if (mCurrentStyle == 1) // ball & stick { // activate "ball & stick" style mMolecules[mCurrentMolecule]->setMoleculeStyle(vl::MS_BallAndStick); // colorize the atoms by their CPK color mMolecules[mCurrentMolecule]->setCPKAtomColors(); // set all the atom radii to 0.30A mMolecules[mCurrentMolecule]->setAtomRadii(0.30f); // set all the bond radii to 0.15A mMolecules[mCurrentMolecule]->setBondRadii(0.15f); } else if (mCurrentStyle == 2) // sticks { // activate "sticks" style mMolecules[mCurrentMolecule]->setMoleculeStyle(vl::MS_Sticks); // colorize the atoms by their CPK color mMolecules[mCurrentMolecule]->setCPKAtomColors(); // set all the bond radii to 0.10A mMolecules[mCurrentMolecule]->setBondRadii(0.10f); } else if (mCurrentStyle == 3) // cpk space fill { // activates "atoms only" style mMolecules[mCurrentMolecule]->setMoleculeStyle(vl::MS_AtomsOnly); // colorize the atoms by their CPK color mMolecules[mCurrentMolecule]->setCPKAtomColors(); // set all the atom radii to their van der Waals radii value as returned by atomInfo(). mMolecules[mCurrentMolecule]->setVanDerWaalsAtomRadii(); } /* SETUP TO RENDER THE ATOM LABELS: ... choose the style: font, color, alignment etc. mMolecules[mCurrentMolecule]->atomLabelTemplate()->setFont( vl::defFontManager()->acquireFont("/font/bitstream-vera/VeraMono.ttf", 10) ); mMolecules[mCurrentMolecule]->atomLabelTemplate()->setColor(vl::white); mMolecules[mCurrentMolecule]->atomLabelTemplate()->setOutlineColor(vl::black); mMolecules[mCurrentMolecule]->atomLabelTemplate()->setOutlineEnabled(true); mMolecules[mCurrentMolecule]->atomLabelTemplate()->setAlignment(vl::AlignHCenter|vl::AlignVCenter); ... enable the atom label rendering at the molecule level mMolecules[mCurrentMolecule]->setShowAtomNames(true); ... enable the atom label rendering at the atom level mMolecules[mCurrentMolecule]->atom(4)->setShowAtomName(true); OTHER COMMON OPERATIONS: ... aromatic ring settings mMolecules[mCurrentMolecule]->setAromaticRingColor(vl::red); mMolecules[mCurrentMolecule]->setAromaticBondsColor(vl::gold); ... geometrical detail for bonds and atoms mMolecules[mCurrentMolecule]->setBondDetail(50); mMolecules[mCurrentMolecule]->setAtomDetail(3); ... toggle visibility by atom type mMolecules[mCurrentMolecule]->setAtomTypeVisible(vl::AT_Hydrogen, false); ... define per-atom color mMolecules[mCurrentMolecule]->atom(4)->setColor(vl::fuchsia); ... define per-atom and per-bond visibility mMolecules[mCurrentMolecule]->atom(5)->setVisible(false); mMolecules[mCurrentMolecule]->bond(6)->setVisible(false); */ /* generates the actual geometry to be rendered, the result of this function will be in mMolecules[mCurrentMolecule]->actorTree() and mMolecules[mCurrentMolecule]->transformTree() */ mMolecules[mCurrentMolecule]->prepareForRendering(); /* remove any previously installed child node */ sceneManager()->tree()->eraseAllChildren(); /* add the node containing the molecule's Actors to the scene */ sceneManager()->tree()->addChild( mMolecules[mCurrentMolecule]->actorTree() ); /* updates the text on top of the window with the current molecule name and style */ updateText(); } /* initialization */ void initEvent() { vl::Log::notify(appletInfo()); /* initialize the text actor */ mText->setText("Drop a MOL2 file inside the window."); mText->setFont( vl::defFontManager()->acquireFont("/font/bitstream-vera/VeraMono.ttf", 10) ); mText->setAlignment( vl::AlignHCenter | vl::AlignTop ); mText->setViewportAlignment( vl::AlignHCenter | vl::AlignTop ); mText->setTextAlignment(vl::TextAlignCenter); mText->translate(0,-5,0); mText->setColor(vl::white); vl::ref<vl::Effect> effect = new vl::Effect; effect->shader()->enable(vl::EN_BLEND); sceneManager()->tree()->addActor(mText.get(), effect.get()); loadMolecule("/mol/molecule.mol2"); } /* Loads the specified mol2 file. */ void loadMolecule(const vl::String& mol2_file) { mCurrentMolecule = 0; vl::loadMOL2( mol2_file, mMolecules ); if (!mMolecules.empty()) updateMolecule(); /* adjust the camera position to nicely see the scene, it also position the rotation pivot to the center of the molecule */ trackball()->adjustView( rendering()->as<vl::Rendering>(), vl::vec3(0,0,1), vl::vec3(0,1,0), 1.0f ); for(size_t i=0; i<mMolecules.size(); ++i) { vl::String msg; msg = "New molecule: " + mMolecules[i]->moleculeName() + " - " + vl::String::fromInt(mMolecules[i]->atomCount()) + " atoms\n"; vl::Log::print(msg); } } /* loads a MOL2 file when it is dropped in the window */ void fileDroppedEvent(const std::vector<vl::String>& files) { /*loads only the first .mol2 file if more are dropped*/ loadMolecule( files[0] ); } /* user controls to change the molecule (if we loaded a multi-MOL2 file) and the style */ void keyPressEvent(unsigned short ch, vl::EKey key) { BaseDemo::keyPressEvent(ch,key); if (key == vl::Key_Up || key == vl::Key_Down || key == vl::Key_Left || key == vl::Key_Right) { if (key == vl::Key_Up ) mCurrentStyle++; if (key == vl::Key_Down) mCurrentStyle--; if (key == vl::Key_Left) mCurrentMolecule--; if (key == vl::Key_Right) mCurrentMolecule++; if (mCurrentMolecule<0) mCurrentMolecule = (int)mMolecules.size()-1; if (mCurrentMolecule>(int)mMolecules.size()-1) mCurrentMolecule = 0; if (mCurrentStyle<0) mCurrentStyle = 3; if (mCurrentStyle>3) mCurrentStyle = 0; updateMolecule(); } } /* updates the text on top of the window with the current molecule name and style */ void updateText() { vl::String msg = mMolecules[mCurrentMolecule]->moleculeName(); msg += vl::Say(" (%n/%n)") << mCurrentMolecule+1 << mMolecules.size(); if (mCurrentStyle == 0) msg += " - Wireframe"; if (mCurrentStyle == 1) msg += " - Ball & Stick"; if (mCurrentStyle == 2) msg += " - Sticks"; if (mCurrentStyle == 3) msg += " - CPK"; msg += "\nuse the arrow keys to change molecule and style"; mText->setText(msg); } protected: std::vector< vl::ref<vl::Molecule> > mMolecules; int mCurrentMolecule; int mCurrentStyle; vl::ref<vl::Text> mText; }; // Have fun!